StepOnePlus Uniformity Test
Rationale
Testing the Baker Lab StepOnePlus machine for any potential variation among wells due to machine error/variability. Machine was calibrated (spatial, background, and dye) on 03/06/2017. To test for any variability, I will run a plate with 96 technical replicates, i.e., the same reaction in each well, that includes amplification of both C and D actin loci (VIC and FAM dyes, respectively.
Methods
- Prepare DNA template: (210 µL total)
- 10 µL Mc16-47t0 (Rivah’s sample, contains clade C)
- 10 µL 942-1 (see previous entry, contains clade D)
- 180 µL H2O
- Prepare qPCR MasterMix:
- Each reaction: 5 µL GTMM; 0.5 µL each of CFor, CRev, CProbe(VIC), DFor, DRev, DProbe(FAM); (no water)
- MasterMix (n=98): 490 µL GTMM; 49 µL each oligo
- Add 8 µL MasterMix to each well
- Add 2 µL mixed C+D DNA template to each well
Results
Raw data files:
library(reshape2)
df <- read.delim("20170307_RC_UniformityTest_data.txt", skip=8)
df <- dcast(df, formula=Well~Target.Name, value.var="Cт")Visualize plates:
library(platetools)
raw_map(data=df$C, well=df$Well, plate=96)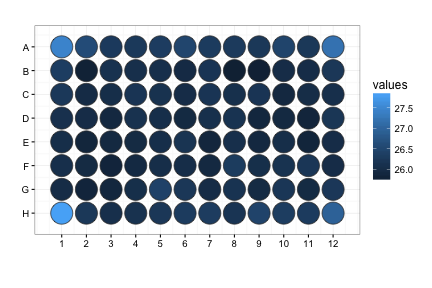
raw_map(data=df$D, well=df$Well, plate=96)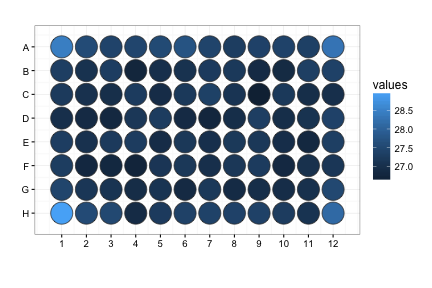
Run some stats:
library(effects)
library(lsmeans)## Loading required package: estimability## Loading required package: methodsdf$row <- factor(substr(df$Well, 1,1))
df$col <- factor(as.numeric(substr(df$Well, 2,3)))
df$corner <- factor(ifelse(df$Well %in% c("A1", "A12", "H1", "H12"), "corner", "not_corner"))
df$border <- factor(ifelse(df$row %in% c("A", "H") | df$col %in% c(1, 12), "border", "not_border"))
str(df)## 'data.frame': 96 obs. of 7 variables:
## $ Well : Factor w/ 96 levels "A1","A10","A11",..: 1 2 3 4 5 6 7 8 9 10 ...
## $ C : num 27.4 26.5 26.2 27.2 26.6 ...
## $ D : num 28.4 27.4 27.4 28.3 27.6 ...
## $ row : Factor w/ 8 levels "A","B","C","D",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ col : Factor w/ 12 levels "1","2","3","4",..: 1 10 11 12 2 3 4 5 6 7 ...
## $ corner: Factor w/ 2 levels "corner","not_corner": 1 2 2 1 2 2 2 2 2 2 ...
## $ border: Factor w/ 2 levels "border","not_border": 1 1 1 1 1 1 1 1 1 1 ...# Are corners different?
mod <- lm(C ~ corner, data=df)
pairs(lsmeans(mod, specs="corner"))## contrast estimate SE df t.ratio p.value
## corner - not_corner 1.2511 0.09363602 94 13.361 <.0001plot(effect("corner", mod), main="Corner effect - C assay")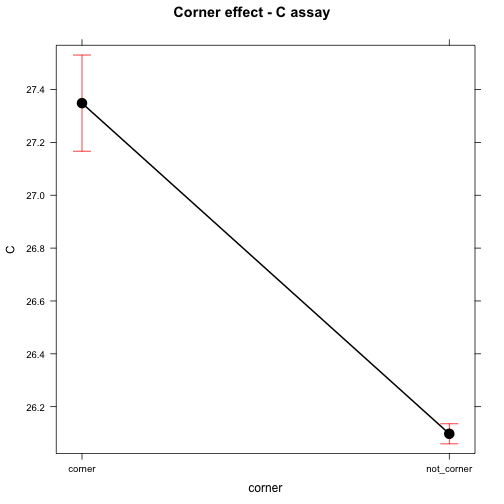
mod <- lm(D ~ corner, data=df)
pairs(lsmeans(mod, specs="corner"))## contrast estimate SE df t.ratio p.value
## corner - not_corner 1.297778 0.1189481 94 10.91 <.0001plot(effect("corner", mod), main="Corner effect - D assay")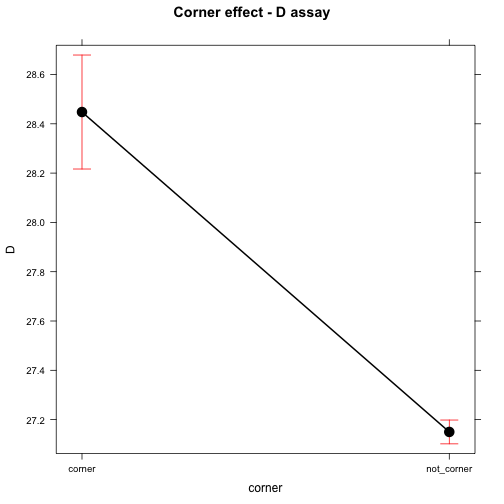
# Are columns different? (excluding corners)
mod <- lm(C ~ col, data=subset(df, corner=="not_corner"))
anova(mod)## Analysis of Variance Table
##
## Response: C
## Df Sum Sq Mean Sq F value Pr(>F)
## col 11 0.19389 0.017626 0.5508 0.8623
## Residuals 80 2.56001 0.032000plot(effect("col", mod), main="Column effect - C assay (excluding corners)")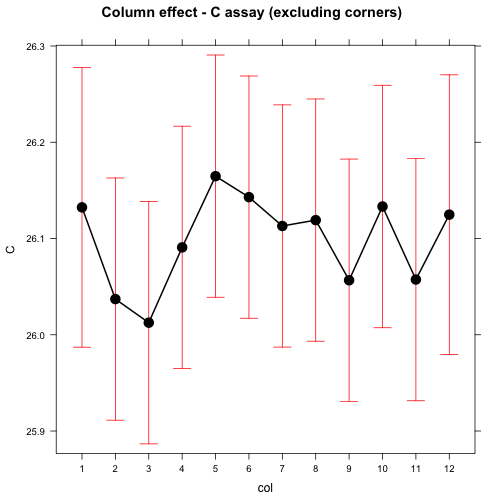
mod <- lm(D ~ col, data=subset(df, corner=="not_corner"))
anova(mod)## Analysis of Variance Table
##
## Response: D
## Df Sum Sq Mean Sq F value Pr(>F)
## col 11 0.3927 0.035699 0.66 0.7716
## Residuals 80 4.3274 0.054093plot(effect("col", mod), main="Column effect - D assay (excluding corners)")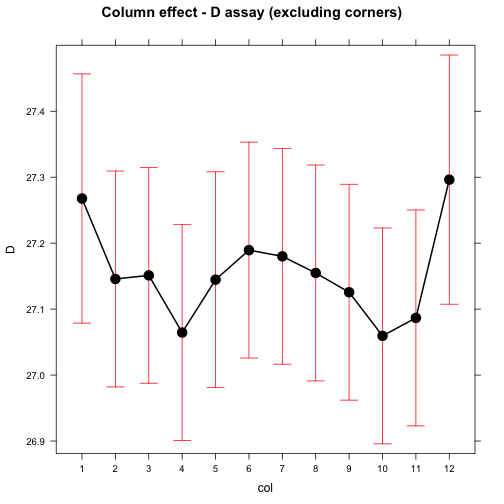
# Are rows different? (excluding corners)
mod <- lm(C ~ row, data=subset(df, corner=="not_corner"))
cld(lsmeans(mod, specs="row"))## row lsmean SE df lower.CL upper.CL .group
## E 25.97977 0.03871915 84 25.90277 26.05676 1
## B 26.02156 0.03871915 84 25.94456 26.09856 1
## D 26.02179 0.03871915 84 25.94479 26.09879 1
## F 26.04711 0.03871915 84 25.97011 26.12411 1
## G 26.08002 0.03871915 84 26.00302 26.15701 12
## C 26.08852 0.03871915 84 26.01152 26.16551 12
## H 26.25853 0.04241470 84 26.17418 26.34288 23
## A 26.35127 0.04241470 84 26.26692 26.43562 3
##
## Confidence level used: 0.95
## P value adjustment: tukey method for comparing a family of 8 estimates
## significance level used: alpha = 0.05plot(effect("row", mod), main="Row effect - C assay (excluding corners)")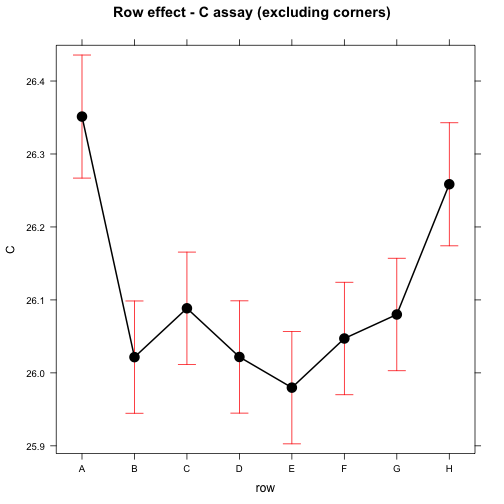
mod <- lm(D ~ row, data=subset(df, corner=="not_corner"))
cld(lsmeans(mod, specs="row"))## row lsmean SE df lower.CL upper.CL .group
## F 27.03702 0.05507012 84 26.92751 27.14654 1
## D 27.04307 0.05507012 84 26.93356 27.15259 1
## C 27.07713 0.05507012 84 26.96762 27.18665 12
## G 27.09902 0.05507012 84 26.98951 27.20854 12
## B 27.12032 0.05507012 84 27.01081 27.22984 12
## E 27.12401 0.05507012 84 27.01450 27.23352 12
## H 27.31521 0.06032630 84 27.19524 27.43518 23
## A 27.46405 0.06032630 84 27.34408 27.58402 3
##
## Confidence level used: 0.95
## P value adjustment: tukey method for comparing a family of 8 estimates
## significance level used: alpha = 0.05plot(effect("row", mod), main="Row effect - D assay (excluding corners)")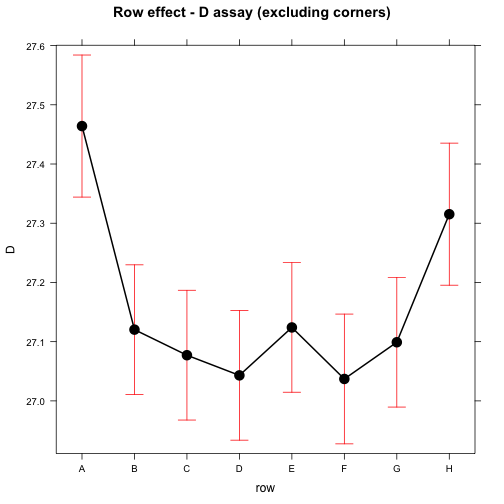
Conclusions:
Corners amplify ~1.3 cycles later than other wells. Other wells in rows A and H (excluding corners!) amplify ~0.3 cycles later than other wells. There is no effect of column.