Porites DNA amplification efficiency
Rationale: Genomic DNA extracted from Porites corals often contains PCR inhibitors, which precludes reliable quantification using qPCR. We are testing ways of cleaning up this DNA so it is suitable for assaying symbiont to host cell ratios with qPCR. Raphael has cleaned 8 Porites DNA samples using Ampure beads, which I am using to create dilution series to run standard curves to calculate amplification efficiency. If amplification efficiency is near 100%, then Ampure beads produce sufficiently clean DNA samples for qPCR.
Samples: 27, 28, 35, 36, 41, 42, 49, 50
Primers: PL1556PET, PL1556r (microsatellite primers from Concepcion et al. 2010, amplifying a 164bp fragment, melting temp. ~ 59°C). Obtained primer stocks from Zac Forsman, diluted aliquot to 10 µM in water.
Each reaction: 6.25 µL SYBR Green MasterMix, 0.5 µL forward primer @ 10 µM, 0.5 µL reverse primer @ 10 µM, 4.25 µL water, 1 µL DNA template
Template: Tenfold dilution series of each sample, five levels
Run: Default settings on StepOnePlus, annealing temperature 59°C
Results:
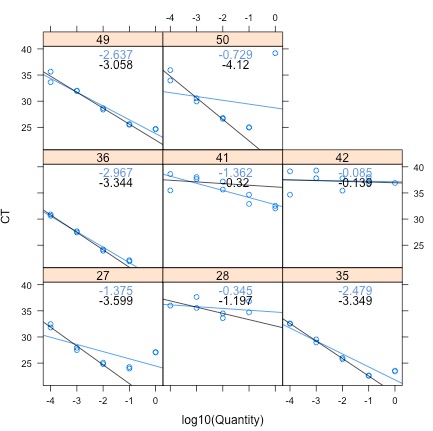
Each panel represents a sample. Blue regression lines and slopes are for all data, black regression lines and slopes are for only 1:100 dilutions and below. Without dilution, all samples are inhibitied, evidenced by slopes greater than -3.3. Once diluted 1:100 and below, 5 samples show passable results without inhibition: 49, 50, 36, 27, and 35. Three samples showed major inhibition at all dilution levels: 41, 42, and 28.
Conclusion: Ampure bead cleaning is not sufficient to produce reliable, inhibitor free DNA from Porites samples.